
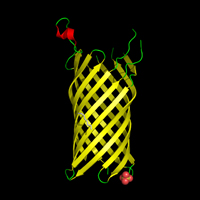
Outer Membrane Proteins >> Outer membrane adhesin OpcA
NAME:Outer membrane adhesin OpcA
PDB IDs: 2VDF
SOURCE ORGANISM: Neisseria menningitidis
SEQUENCE: UniProt Q9AE79>>
ORIENTATION IN MEMBRANE: OPM >>
CRYSTALLIZATION CONDITIONS:
| LCP 200 nL | Ppt solution 2 µL |
| Lipid 60 %w/w | Protein Solution 40 %w/w | |
| Lipid: 100 %w/w Monoolein | Protein: 12 mg/mL OpcA | Buffer: 50 mM Hepes pH 7.0 |
| Buffer: 50 mM Bis tris propane pH 7.0 | Precipitant: 18 %v/v PEG 400 | |
| Detergent: 0.1 %w/v DDM | Salt: 0.1 M K sulfate |
Temperature (°C): 20
Crystal growth time (d): 20 – 30 Crystal size (µm): 25 x 25 x 70
Reference: V. Cherezov, W. Liu, J. P. Derrick, B. Luan, A. Aksimentiev, V. Katritch and M. Caffrey. 2008. In meso crystal structure and docking simulations suggest an alternative proteoglycan binding site in the OpcA outer membrane adhesin. Proteins 71: 24-34 >>
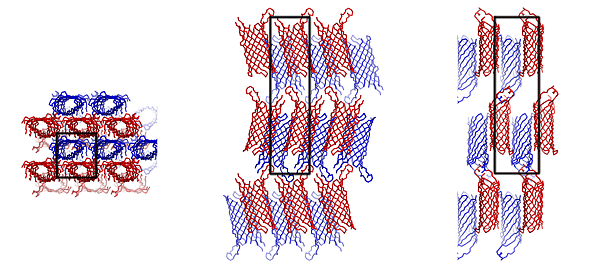
SPACE GROUP: P 21 21 21 UNIT CELL: 37.86 42.50 150.30 90.0 90.0 90.0
CRYSTAL PACKING:
CONTACT US: USC | Cherezov Lab | cherezov@usc.edu